165-在linux中用脚本探索基因组区域
刘小泽写于2020.2.12 看到一个生信前辈DaveTang的博客中一篇很久以前的博文,虽然已经过去了5年时间,但其中的思想还是值得学习的。今天的内容就是:https://davetang.org/muse/2013/01/18/defining-genomic-regions/,使用脚本和GTF文件获取外显子、内含子、基因间区信息
前言
DaveTang的这篇博客更新于2014年,那时转录组测序很火热。RNA-Seq往往是把reads比对回基因组或转录组,然后就是对reads进行注释,看看它们落在什么位置,其实定量也是其中一步。这里,作者就带我们看看他是如何获得外显子(exonic)、内含子(intronic)、基因间区(intergenic regions)的坐标的。
首先下载参考基因组注释
# hg38版本
wget -c ftp://ftp.ebi.ac.uk/pub/databases/gencode/Gencode_human/release_33/gencode.v33.annotation.gtf.gz
zcat gencode.v33.annotation.gtf.gz | head -n6
##description: evidence-based annotation of the human genome (GRCh38), version 33 (Ensembl 99)
##provider: GENCODE
##contact: gencode-help@ebi.ac.uk
##format: gtf
##date: 2019-12-13
chr1 HAVANA gene 11869 14409 . + . gene_id "ENSG00000223972.5"; gene_type "transcribed_unprocessed_pseudogene"; gene_name "DDX11L1"; level 2; hgnc_id "HGNC:37102"; havana_gene "OTTHUMG00000000961.2";
其中的第三列说明了feature的类型,其中会有exon、CDS、UTR等等
zcat gencode.v33.annotation.gtf.gz| grep -v "^#" | cut -f3 | sort | uniq -c | sort -k1rn
1377112 exon
763824 CDS
311868 UTR
227912 transcript
87848 start_codon
80187 stop_codon
60662 gene
119 Selenocysteine
它们之间的关系,可以看看:http://blog.sciencenet.cn/blog-1271266-792469.html以及http://www.dxy.cn/bbs/topic/36728037
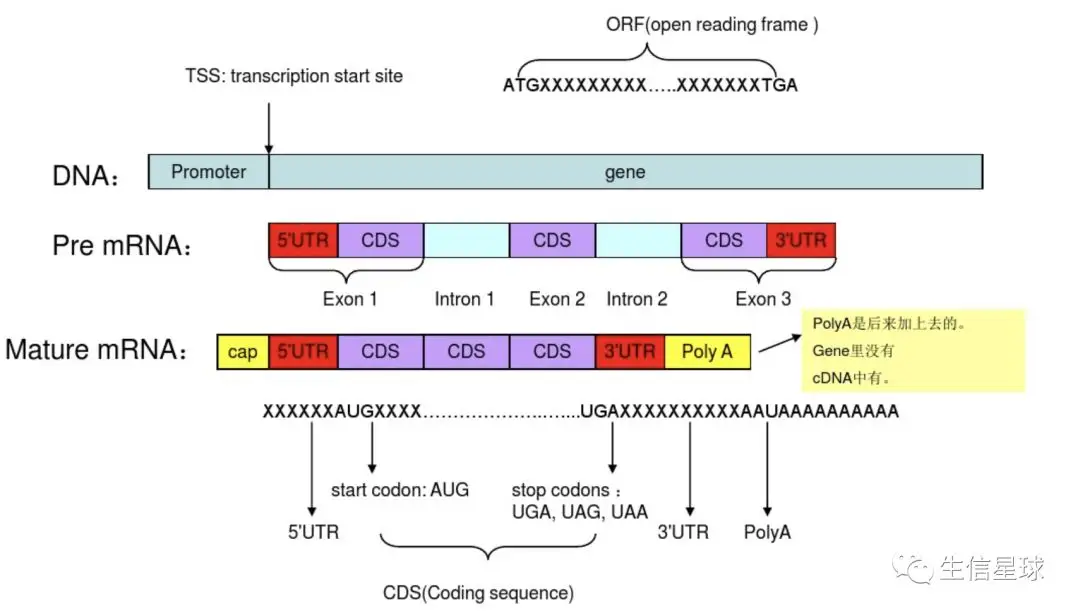
**UTR(UntranslatedRegions)**即非翻译区,是信使RNA(mRNA)分子两端的非编码片段,UTR在DNA序列中算是外显子;
5’-UTR从mRNA起点的甲基化鸟嘌呤核苷酸帽延伸至AUG起始密码子,3’-UTR从编码区末端的终止密码子延伸至多聚A尾巴(Poly-A)的末端;
基因间区是每个基因之间的间隔序列,不属于外显子也不属于内含子,它是non coding;
【图片中描述的过程:基因经过转录形成 pre-mRNA,这里面包含着内含子和外显子(5端是一个外显子,但是这段外显子不仅包含CDS,还包含5’ UTR;3端也是以外显子结束,但它也是不仅包含CDS,还包含3’ UTR),经过剪接形成成熟mRNA,内含子已减掉。如果抛开后来加上去的cap和poly A的话,这时全是外显子,但是不全是CDS,因为只有中间的那部分以起始密码子开始、以终止密码子结束的片段才是CDS,只有这部分才会被翻译成蛋白质】
**转录本(transcript)**是基因通过转录形成的一种或多种可供编码蛋白质的成熟的mRNA。一个基因由于可变剪切现象,可能产生多个转录本。【基因转录的过程,首先是形成前体mRNA,通过剪切内含子连接外显子,5’端加帽及3’端加尾之后形成成熟的mRNA。可变剪切就是:在剪切的过程中可能会剪切掉外显子,也有可能保留部分内含子,因此会形成多种mRNA即多个转录本】,而我们平时研究某个基因的功能,实际是研究它的一个转录本编码的蛋白的功能。一般情况下它的不同的转录本分布在不同类型的细胞中,当然也有可能多种转录本同时存在于某一细胞中。
编码区(Coding region,CDS) 是mRNA序列中编码蛋白质的那部分序列
获得外显子、内含子以及基因间区
外显子的获得 => mergeBed
从GTF文件中其实已经能找到exon的踪迹,但是同一个基因下的各个exon可能会有交集,于是这里作者在统计时,将存在重叠的外显子融合在一起(这里只是采用最简单的方法进行探索,但不是最准确的)
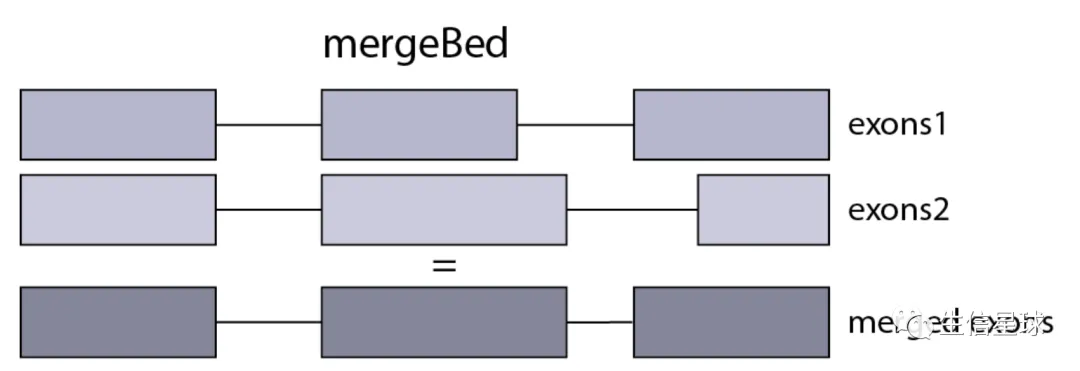
使用bedtools的mergeBed功能,但前提是先使用sortBed将坐标进行排序
# 首先挑出exon的起始和终止位点
zcat gencode.v33.annotation.gtf.gz | awk 'BEGIN{OFS="\t";} $3=="exon" {print $1,$4-1,$5}' | head -n5
chr1 11868 12227
chr1 12612 12721
chr1 13220 14409
chr1 12009 12057
chr1 12178 12227
# 然后进行排序,接着进行merge
zcat gencode.v33.annotation.gtf.gz | awk 'BEGIN{OFS="\t";} $3=="exon" {print $1,$4-1,$5}' | sortBed | mergeBed -i - | gzip >gencode.v33.exon.annotation.gtf.gz
zcat gencode.v33.exon.annotation.gtf.gz | head -n5
chr1 11868 12227
chr1 12612 12721
chr1 12974 13052
chr1 13220 14501
chr1 15004 15038
小tip: 上面👆的
awk命令中出现了一个{print $1,$4-1,$5},其中$4是指的GTF的第4列,即起始位点。它要减一是因为要从GTF变为BED,而它们的坐标系统不一致:GTF has 1-based start coordinates, and BED has 0-based start coordinates
内含子的获得 => subtractBed
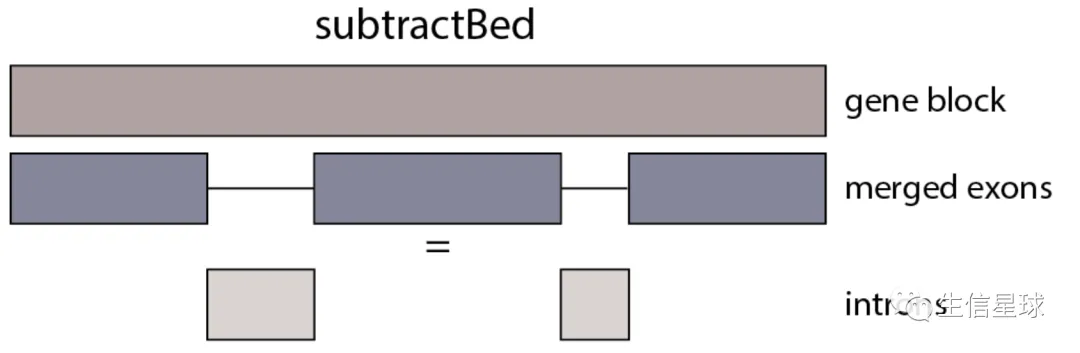
# 选择GTF中gene的feature,然后排除merged exon
zcat gencode.v33.annotation.gtf.gz | awk 'BEGIN{OFS="\t";} $3=="gene" {print $1,$4-1,$5}' | sortBed | subtractBed -a stdin -b gencode.v33.exon.annotation.gtf.gz | gzip >gencode.v33.intron.annotation.gtf.gz
zcat gencode.v33.intron.annotation.gtf.gz | head -n5
# 结果可以和上面👆的exon结果对照
chr1 12227 12612
chr1 12721 12974
chr1 13052 13220
chr1 14501 15004
chr1 15038 15795
基因间区的获得 => complementBed
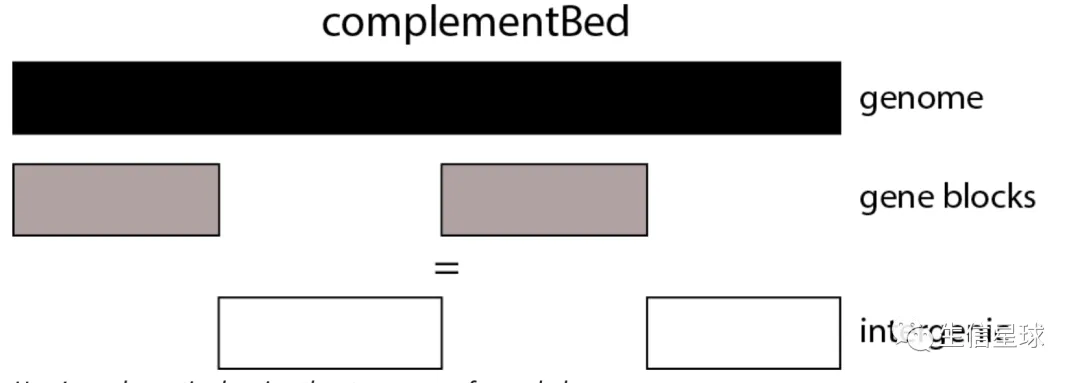
需要先获得整个基因组各个染色体的长度,然后从中排除掉gene
# 各个染色体的长度(在bedtools complementBed的帮助文档中)
mysql --user=genome --host=genome-mysql.cse.ucsc.edu -A -e "select chrom, size from hg38.chromInfo" >hg38.genome
# 去掉hg38.genome第一行注释:chrom size
sed -i '1d' hg38.genome
# 排除gene
zcat gencode.v33.annotation.gtf.gz | awk 'BEGIN{OFS="\t";} $3=="gene" {print $1,$4-1,$5}' | sortBed | complementBed -i stdin -g hg38.genome | gzip >gencode.v33.intergenic.annotation.gtf.gz
但这里报了个错:
Error: Sorted input specified, but the file stdin has the following record with a different sort order than the genomeFile hg38.genome
实际上问题就是因为我们的sort bed后的chrxx不是按照数字排序的(http://asearchforsolutions.blogspot.com/2018/11/error-sorted-input-specified-but-file.html)
zcat gencode.v33.annotation.gtf.gz | awk 'BEGIN{OFS="\t";} $3=="gene" {print $1,$4-1,$5}' | sortBed | cut -f1 | uniq
chr1
chr10
chr11
chr12
chr13
chr14
chr15
chr16
chr17
chr18
# 而这里的hg38.genome却是拍好顺序的
cat hg38.genome | cut -f1 | uniq | head
chr1
chr2
chr3
chr4
chr5
chr6
chr7
chrX
chr8
chr9
# 因此我们“恃强凌弱”,将hg38.genome改成和sortBed后一致的(相比之下hg38.genome更容易修改),也不按照字母排序
cat hg38.genome| sort -k1.4 >hg38-2.genome
head hg38-2.genome
chr1
chr10
chr10_GL383545v1_alt
chr10_GL383546v1_alt
chr10_KI270824v1_alt
chr10_KI270825v1_alt
chr10_KN196480v1_fix
chr10_KN538365v1_fix
chr10_KN538366v1_fix
chr10_KN538367v1_fix
于是,继续进行代码,就没有问题啦:
zcat gencode.v33.annotation.gtf.gz | awk 'BEGIN{OFS="\t";} $3=="gene" {print $1,$4-1,$5}' | sortBed | complementBed -i stdin -g hg38-2.genome | gzip >gencode.v33.intergenic.annotation.gtf.gz
zcat gencode.v33.intergenic.annotation.gtf.gz | head -n5
chr1 0 11868
chr1 31109 34553
chr1 36081 52472
chr1 53312 57597
chr1 64116 65418
现在三个文件具备了,就可以进行探索了
首先看基因组上外显子、内含子、基因间区各占多少比例呢?
cat >coverage.pl ,然后输入下面的代码
#!/bin/env perl
use strict;
use warnings;
my $exon_file = "gencode.v33.exon.annotation.gtf.gz";
my $intergenic_file = "gencode.v33.intergenic.annotation.gtf.gz";
my $intron_file = "gencode.v33.intron.annotation.gtf.gz";
my $exon_coverage = coverage($exon_file);
my $intergenic_coverage = coverage($intergenic_file);
my $intron_coverage = coverage($intron_file);
my $total = $exon_coverage + $intergenic_coverage + $intron_coverage;
printf "Exon: %.2f\n", $exon_coverage*100/$total;
printf "Intron: %.2f\n", $intron_coverage*100/$total;
printf "Intergenic: %.2f\n", $intergenic_coverage*100/$total;
sub coverage {
my ($infile) = @_;
my $coverage = 0;
open(IN,'-|',"zcat $infile") || die "Could not open $infile: $!\n";
while(<IN>){
chomp;
my ($chr, $start, $end) = split(/\t/);
my $c = $end - $start;
$coverage += $c;
}
close(IN);
return($coverage);
}
exit(0);
最后看到内含子>间区>外显子:
# perl coverage.pl
Exon: 4.25
Intron: 51.98
Intergenic: 43.77
关于非翻译区untranslated region (UTR)
以转录本ENST00000335295为例:
zcat gencode.v33.annotation.gtf.gz | grep ENST00000335295
chr11 HAVANA transcript 5225464 5227071 . - . gene_id "ENSG00000244734.4"; transcript_id "ENST00000335295.4"; gene_type "protein_coding"; gene_name "HBB"; transcript_type "protein_coding"; transcript_name "HBB-201"; level 2; protein_id "ENSP00000333994.3"; transcript_support_level "1"; hgnc_id "HGNC:4827"; tag "CAGE_supported_TSS"; tag "basic"; tag "MANE_Select"; tag "appris_principal_1"; tag "CCDS"; ccdsid "CCDS7753.1"; havana_gene "OTTHUMG00000066678.8"; havana_transcript "OTTHUMT00000495006.2";
chr11 HAVANA exon 5226930 5227071 . - . gene_id "ENSG00000244734.4"; transcript_id "ENST00000335295.4"; gene_type "protein_coding"; gene_name "HBB"; transcript_type "protein_coding"; transcript_name "HBB-201"; exon_number 1; exon_id "ENSE00001829867.2"; level 2; protein_id "ENSP00000333994.3"; transcript_support_level "1"; hgnc_id "HGNC:4827"; tag "CAGE_supported_TSS"; tag "basic"; tag "MANE_Select"; tag "appris_principal_1"; tag "CCDS"; ccdsid "CCDS7753.1"; havana_gene "OTTHUMG00000066678.8"; havana_transcript "OTTHUMT00000495006.2";
chr11 HAVANA CDS 5226930 5227021 . - 0 gene_id "ENSG00000244734.4"; transcript_id "ENST00000335295.4"; gene_type "protein_coding"; gene_name "HBB"; transcript_type "protein_coding"; transcript_name "HBB-201"; exon_number 1; exon_id "ENSE00001829867.2"; level 2; protein_id "ENSP00000333994.3"; transcript_support_level "1"; hgnc_id "HGNC:4827"; tag "CAGE_supported_TSS"; tag "basic"; tag "MANE_Select"; tag "appris_principal_1"; tag "CCDS"; ccdsid "CCDS7753.1"; havana_gene "OTTHUMG00000066678.8"; havana_transcript "OTTHUMT00000495006.2";
chr11 HAVANA start_codon 5227019 5227021 . - 0 gene_id "ENSG00000244734.4"; transcript_id "ENST00000335295.4"; gene_type "protein_coding"; gene_name "HBB"; transcript_type "protein_coding"; transcript_name "HBB-201"; exon_number 1; exon_id "ENSE00001829867.2"; level 2; protein_id "ENSP00000333994.3"; transcript_support_level "1"; hgnc_id "HGNC:4827"; tag "CAGE_supported_TSS"; tag "basic"; tag "MANE_Select"; tag "appris_principal_1"; tag "CCDS"; ccdsid "CCDS7753.1"; havana_gene "OTTHUMG00000066678.8"; havana_transcript "OTTHUMT00000495006.2";
chr11 HAVANA exon 5226577 5226799 . - . gene_id "ENSG00000244734.4"; transcript_id "ENST00000335295.4"; gene_type "protein_coding"; gene_name "HBB"; transcript_type "protein_coding"; transcript_name "HBB-201"; exon_number 2; exon_id "ENSE00001057381.1"; level 2; protein_id "ENSP00000333994.3"; transcript_support_level "1"; hgnc_id "HGNC:4827"; tag "CAGE_supported_TSS"; tag "basic"; tag "MANE_Select"; tag "appris_principal_1"; tag "CCDS"; ccdsid "CCDS7753.1"; havana_gene "OTTHUMG00000066678.8"; havana_transcript "OTTHUMT00000495006.2";
chr11 HAVANA CDS 5226577 5226799 . - 1 gene_id "ENSG00000244734.4"; transcript_id "ENST00000335295.4"; gene_type "protein_coding"; gene_name "HBB"; transcript_type "protein_coding"; transcript_name "HBB-201"; exon_number 2; exon_id "ENSE00001057381.1"; level 2; protein_id "ENSP00000333994.3"; transcript_support_level "1"; hgnc_id "HGNC:4827"; tag "CAGE_supported_TSS"; tag "basic"; tag "MANE_Select"; tag "appris_principal_1"; tag "CCDS"; ccdsid "CCDS7753.1"; havana_gene "OTTHUMG00000066678.8"; havana_transcript "OTTHUMT00000495006.2";
chr11 HAVANA exon 5225464 5225726 . - . gene_id "ENSG00000244734.4"; transcript_id "ENST00000335295.4"; gene_type "protein_coding"; gene_name "HBB"; transcript_type "protein_coding"; transcript_name "HBB-201"; exon_number 3; exon_id "ENSE00001600613.2"; level 2; protein_id "ENSP00000333994.3"; transcript_support_level "1"; hgnc_id "HGNC:4827"; tag "CAGE_supported_TSS"; tag "basic"; tag "MANE_Select"; tag "appris_principal_1"; tag "CCDS"; ccdsid "CCDS7753.1"; havana_gene "OTTHUMG00000066678.8"; havana_transcript "OTTHUMT00000495006.2";
chr11 HAVANA CDS 5225601 5225726 . - 0 gene_id "ENSG00000244734.4"; transcript_id "ENST00000335295.4"; gene_type "protein_coding"; gene_name "HBB"; transcript_type "protein_coding"; transcript_name "HBB-201"; exon_number 3; exon_id "ENSE00001600613.2"; level 2; protein_id "ENSP00000333994.3"; transcript_support_level "1"; hgnc_id "HGNC:4827"; tag "CAGE_supported_TSS"; tag "basic"; tag "MANE_Select"; tag "appris_principal_1"; tag "CCDS"; ccdsid "CCDS7753.1"; havana_gene "OTTHUMG00000066678.8"; havana_transcript "OTTHUMT00000495006.2";
chr11 HAVANA stop_codon 5225598 5225600 . - 0 gene_id "ENSG00000244734.4"; transcript_id "ENST00000335295.4"; gene_type "protein_coding"; gene_name "HBB"; transcript_type "protein_coding"; transcript_name "HBB-201"; exon_number 3; exon_id "ENSE00001600613.2"; level 2; protein_id "ENSP00000333994.3"; transcript_support_level "1"; hgnc_id "HGNC:4827"; tag "CAGE_supported_TSS"; tag "basic"; tag "MANE_Select"; tag "appris_principal_1"; tag "CCDS"; ccdsid "CCDS7753.1"; havana_gene "OTTHUMG00000066678.8"; havana_transcript "OTTHUMT00000495006.2";
chr11 HAVANA UTR 5227022 5227071 . - . gene_id "ENSG00000244734.4"; transcript_id "ENST00000335295.4"; gene_type "protein_coding"; gene_name "HBB"; transcript_type "protein_coding"; transcript_name "HBB-201"; exon_number 1; exon_id "ENSE00001829867.2"; level 2; protein_id "ENSP00000333994.3"; transcript_support_level "1"; hgnc_id "HGNC:4827"; tag "CAGE_supported_TSS"; tag "basic"; tag "MANE_Select"; tag "appris_principal_1"; tag "CCDS"; ccdsid "CCDS7753.1"; havana_gene "OTTHUMG00000066678.8"; havana_transcript "OTTHUMT00000495006.2";
chr11 HAVANA UTR 5225464 5225600 . - . gene_id "ENSG00000244734.4"; transcript_id "ENST00000335295.4"; gene_type "protein_coding"; gene_name "HBB"; transcript_type "protein_coding"; transcript_name "HBB-201"; exon_number 3; exon_id "ENSE00001600613.2"; level 2; protein_id "ENSP00000333994.3"; transcript_support_level "1"; hgnc_id "HGNC:4827"; tag "CAGE_supported_TSS"; tag "basic"; tag "MANE_Select"; tag "appris_principal_1"; tag "CCDS"; ccdsid "CCDS7753.1"; havana_gene "OTTHUMG00000066678.8"; havana_transcript "OTTHUMT00000495006.2";
这个转录本(HBB)在chr11上,从5227071开始,到5225464为止【因为它位于负链】
另外看到两个UTR:一个从5227071开始到5227022(3’ UTR),一个从5225600开始到5225464(5’ UTR)【一头一尾,正好包括进了整个transcript】
那么问题来了,究竟多少转录本定义了UTR呢?
cat >check_utr.pl,然后输入下面的代码
#!/bin/env perl
use strict;
use warnings;
my $usage = "Usage: $0 <infile.annotation.gtf.gz>\n";
my $infile = shift or die $usage;
if ($infile =~ /\.gz/){
open(IN,'-|',"gunzip -c $infile") || die "Could not open $infile: $!\n";
} else {
open(IN,'<',$infile) || die "Could not open $infile: $!\n";
}
my %transcript = ();
my $current_transcript = '';
while(<IN>){
chomp;
next if (/^#/);
#chr11 HAVANA transcript 65265233 65273940 . + . gene_id "ENSG00000251562.3"; transcript_id "ENST00000534336.1"; gene_type "processed_transcript"; gene_status "KNOWN"; gene_name "MALAT1"; transcript_type "non_coding"; transcript_status "KNOWN"; transcript_name "MALAT1-001"; level 2; havana_gene "OTTHUMG00000166322.1"; havana_transcript "OTTHUMT00000389143.1";
my ($chr,$source,$type,$start,$end,$score,$strand,$phase,$annotation) = split(/\t/);
my @annotation = split(/;\s/,$annotation);
my $transcript_id = 'none';
if ($type eq 'transcript'){
foreach my $blah (@annotation){
my ($type,$name) = split(/\s+/,$blah);
if ($type eq 'transcript_id'){
$current_transcript = $name;
$current_transcript =~ s/"//g;
$transcript{$current_transcript} = 0;
}
}
if ($current_transcript eq 'none'){
die "No name for entry $.\n";
}
}
if ($type eq 'UTR'){
$transcript{$current_transcript}++;
}
}
close(IN);
foreach my $transcript (keys %transcript){
print "$transcript\t$transcript{$transcript}\n";
}
exit(0);
然后
perl check_utr.pl gencode.v33.annotation.gtf.gz | gzip >transcript_utr_number.out.gz
# 检查总共多少转录本
zcat transcript_utr_number.out | wc -l
227912
# 看一下其中包含UTR的转录本数量,看到有130008个转录本中没有定义UTR
zcat transcript_utr_number.out | cut -f2 | sort | uniq -c | sort -k1rn | head
130008 0
36305 2
25151 3
12182 1
10905 4
4817 5
2508 6
1421 7
931 8
683 9
# 发现最多的一个转录本定义了63个UTR
zcat transcript_utr_number.out | awk '$2==63 {print}'
# ENST00000617010.2 63
# 它属于基因:NBPF10
结果发现:约有57%的GENCODE transcripts没有UTR,有36305个transcripts含有常规的2个UTR,25151个transcripts含有3个UTR(可能其中一个UTR被分割开了);最多UTR的当属ENST00000617010.2
问题又来了:寻找 5‘ UTR 与 3’UTR
刚才查找了这么多的UTR,那么它们到底哪个是5‘ UTR,哪个是3’ UTR?这个GENCODE的注释文件中也没有进行说明
这里假定:靠近转录本起始位点的是5‘ ,反之是3’
#!/bin/env perl
use strict;
use warnings;
my $usage = "Usage: $0 <infile.annotation.gtf>\n";
my $infile = shift or die $usage;
if ($infile =~ /\.gz/){
open(IN,'-|',"gunzip -c $infile") || die "Could not open $infile: $!\n";
} else {
open(IN,'<',$infile) || die "Could not open $infile: $!\n";
}
my %transcript = ();
my $current_transcript = '';
my $transcript_start = 0;
my $transcript_end = 0;
while(<IN>){
chomp;
next if (/^#/);
#chr11 HAVANA transcript 65265233 65273940 . + . gene_id "ENSG00000251562.3"; transcript_id "ENST00000534336.1"; gene_type "processed_transcript"; gene_status "KNOWN"; gene_name "MALAT1"; transcript_type "non_coding"; transcript_status "KNOWN"; transcript_name "MALAT1-001"; level 2; havana_gene "OTTHUMG00000166322.1"; havana_transcript "OTTHUMT00000389143.1";
my ($chr,$source,$type,$start,$end,$score,$strand,$phase,$annotation) = split(/\t/);
my @annotation = split(/;\s/,$annotation);
my $transcript_id = 'none';
if ($type eq 'transcript'){
foreach my $blah (@annotation){
my ($type,$name) = split(/\s+/,$blah);
if ($type eq 'transcript_id'){
$current_transcript = $name;
$current_transcript =~ s/"//g;
$transcript_start = $start;
$transcript_end = $end;
}
}
if ($current_transcript eq 'none'){
die "No name for entry $.\n";
}
}
if ($type eq 'UTR'){
my $region = '';
if ($strand eq '+'){
my $dis_to_start = abs($start - $transcript_start);
my $dis_to_end = abs($start - $transcript_end);
$region = $dis_to_start < $dis_to_end ? '5_UTR' : '3_UTR';
} else {
my $dis_to_start = abs($end - $transcript_end);
my $dis_to_end = abs($end - $transcript_start);
$region = $dis_to_start < $dis_to_end ? '5_UTR' : '3_UTR';
}
print join ("\t", $chr, $start, $end, $region, $current_transcript, $strand),"\n";
}
}
close(IN);
exit(0);
接着:
perl print_utr.pl gencode.v33.annotation.gtf.gz | gzip > transcript_utr.bed.gz
# 检查结果
zcat transcript_utr.bed.gz | head -n5
chr1 65419 65433 5_UTR ENST00000641515.2 +
chr1 65520 65564 5_UTR ENST00000641515.2 +
chr1 70006 71585 3_UTR ENST00000641515.2 +
chr1 69055 69090 5_UTR ENST00000335137.4 +
chr1 70006 70108 3_UTR ENST00000335137.4 +
这样,以后即使出了新的UTR注释,那么也能比较距离当前的5‘UTR近还是3’UTR近了
启动子区域
我们可以从GTF中获得每个转录本的起始位点,然后再加某个值就可以得到启动子坐标
#!/bin/env perl
use strict;
use warnings;
my $usage = "Usage: $0 <infile.annotation.gtf> <padding>\n";
my $infile = shift or die $usage;
my $span = shift or die $usage;
if ($span !~ /^\d+$/){
die "Please enter a numeric value for the padding\n";
}
my $hg38 = 'hg38-2.genome';
my %hg38 = ();
open(IN,'<',$hg38) || die "Could not open $hg38: $!\n";
while(<IN>){
chomp;
#chr9_gl000201_random 36148
my ($chr, $end) = split(/\t/);
$hg38{$chr} = $end;
}
close(IN);
if ($infile =~ /\.gz/){
open(IN,'-|',"gunzip -c $infile") || die "Could not open $infile: $!\n";
} else {
open(IN,'<',$infile) || die "Could not open $infile: $!\n";
}
while(<IN>){
chomp;
next if (/^#/);
#chr11 HAVANA transcript 65265233 65273940 . + . gene_id "ENSG00000251562.3"; transcript_id "ENST00000534336.1"; gene_type "processed_transcript"; gene_status "KNOWN"; gene_name "MALAT1"; transcript_type "non_coding"; transcript_status "KNOWN"; transcript_name "MALAT1-001"; level 2; havana_gene "OTTHUMG00000166322.1"; havana_transcript "OTTHUMT00000389143.1";
my ($chr,$source,$type,$start,$end,$score,$strand,$phase,$annotation) = split(/\t/);
next unless $type eq 'transcript';
my @annotation = split(/;\s/,$annotation);
my $transcript_id = 'none';
foreach my $blah (@annotation){
my ($type,$name) = split(/\s+/,$blah);
if ($type eq 'transcript_id'){
$transcript_id = $name;
$transcript_id =~ s/"//g;
}
}
if ($transcript_id eq 'none'){
die "No name for entry $.\n";
}
my $promoter_start = '';
my $promoter_end = '';
if ($strand eq '+'){
$promoter_start = $start - $span;
$promoter_end = $start + $span;
} else {
$promoter_start = $end - $span;
$promoter_end = $end + $span;
}
if ($promoter_start < 0){
warn "Adjusted promoter start to 0\n";
$promoter_start = 0;
} elsif ($promoter_end > $hg38{$chr}){
warn "Adjusted promoter end to $hg38{$chr}\n";
$promoter_end = $hg38{$chr};
}
print join("\t",$chr,$promoter_start,$promoter_end,$transcript_id,0,$strand),"\n";
}
close(IN);
exit(0);
然后,
perl promoter.pl gencode.v33.annotation.gtf.gz 200 | head -n5
chr1 11669 12069 ENST00000456328.2 0 +
chr1 11810 12210 ENST00000450305.2 0 +
chr1 29370 29770 ENST00000488147.1 0 -
chr1 17236 17636 ENST00000619216.1 0 -
chr1 29354 29754 ENST00000473358.1 0 +